Submit Locally, Run Globally
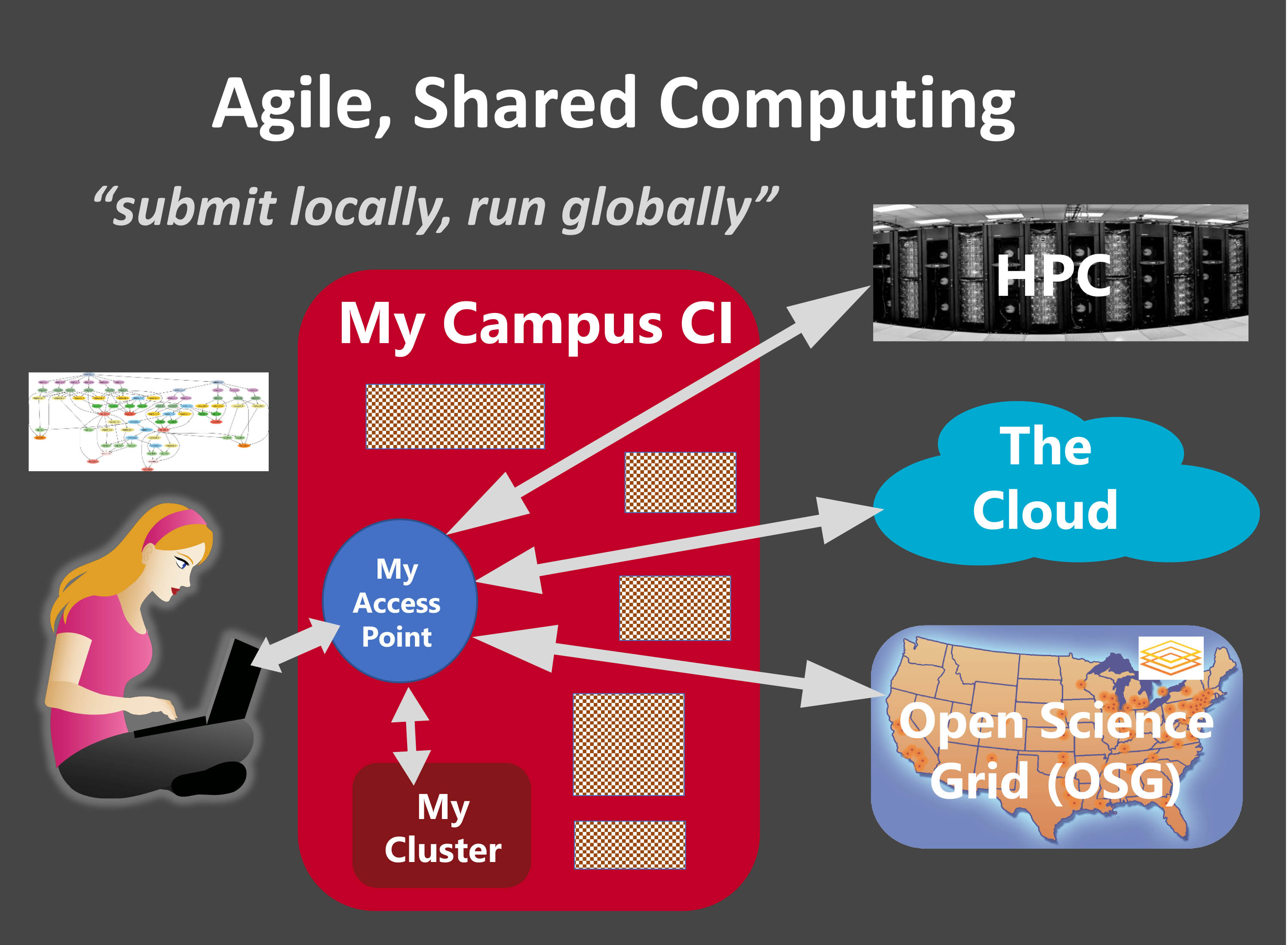
Credit: Miron Livny
The PATh project offers technologies and services that enable researchers to harness through a single interface, and from the comfort of their “home directory”, computing capacity offered by a global and diverse collection of resources.
The OSG offers via the OSG Connect service researchers with an OSG account and an Access Point to the Open Science Pool, a globally deployed HTCondor pool. The capacity offered by this pool comes from more than 130 clusters that are part of the OSG compute federation, and make some of their capacity available to support Open Science. This includes clusters at campuses that are funded under the NSF CC* program.
The The HTCondor Software Suite (HTCSS) offers the technology for these access points that can be deployed and operated by individuals, science collaborations and campuses. Researchers can use such a private/local Access Point to harness the power of locally and remotely deployed HTCondor execution points. This includes institutional and national computing systems (like the Open Science Pool) as well as commercial clouds.
Below are some examples of how researchers enhanced their computational throughput by leveraging the “submit locally, run globally” capabilities offered by PATh Access Points.
AI-Driven Chemistry Dr. Roman Zubatyuk from the group of Prof. Olexandr Isayev at the Carnegie Mellon University chemistry department uses the access point provided by the OSG to develop a supervised machine learning method that more quickly predicts the chemical behavior of small molecules and potential drugs. Via this single access point, he is able to deploy workers globally in support of a high-throughput “Manager-worker” application. Roman reported about his OSG experience in a presentation delivered at the 2020 HTCondor Week workshop.
Astrophysics The IceCube collaboration has operated an HTCSS access point at the Wisconsin IceCube Particle Astrophysics Center for several years. It was used to access GPU capacity offered by the OSG compute federation that includes more than 20 sites of the Pacific Research Platform (PRP). As part of an NSF EAGER project, a team led by OSG Executive Director Frank Wuerthwein and Developer Igor Sfiligoi from the SDSC used this access point to study large-scale bursting of IceCube simulations on resources acquired from commercial cloud. In 2019, the team demonstrated a burst at the Supercomputing 19 conference that harnessed the capacity of 51K GPUs from three cloud providers and scattered across 26 cloud regions, a feat recognized as “the largest cloud simulation in history” in a keynote presentation at the conference.
Bat Genomics Dr. Ariadna Morales, used the Access Point recently deployed at the American Museum of Natural History (AMNH) to harness the capacity of local and OSG capacity in support of her work to analyze the whole genome data of 20 bats. Ariadna, a postdoc at the AMNH, used the local Access Point to run 80K compute jobs in one year. More details of her experience and other AMNH researchers can be found in the AMNH presentation at the 2020 OSG All Hands Meeting.
Gravitational Waves When the LIGO collaboration realized the need for additional capacity to analyze the observed data that led to the Nobel prize-winning detection of gravitational waves, they deployed an HTCSS Access Point that enabled them to expend their High Throughput computing capabilities to include capacity from the OSG federation and XRAC allocations. As an early adopter of HTCSS technologies, the additional capacity provided via the new Access Point easily integrated with the computing and data infrastructure of the collaboration. Deployed more than five years ago, this same access point remains an integral part of the LIGO distributed computing infrastructure, routinely integrating capacity offered by the OSG, international collaborators, XRAC allocations, and the capacity of the LIGO data grid. Read more here.
Drug Screening The Small Molecule Screening Facility (SMSF) within UW-Madison’s Carbone Cancer Center Drug Development Core has been using an access point deployed by the UW-Madison Center for High Throughput Computing (CHTC) to perform computational docking-based screening of potential drug molecules. In just the month of November 2020, the SMSF used almost 2M core hours provided by the OSG for two projects. In collaboration with Prof. Shashank Dravid at Creighton University, one project aims to identify inhibitors of synaptic recruiting proteins for ionotropic glutamate receptors. This novel protein-protein interaction target required the docking of 14 million representations of 8 million molecules from the Aldrich chemical library using a consensus methodology developed by the SMSF to incorporate 6 different docking programs. Another project with Dr. Weiping Tang at UW-Madison leverages the same methodology to develop novel PROTACs, that selectively degrade specific protein targets to regulate biological activity. In order to identify a ligand molecule that can recruit PROTACs to biological targets, the SMSF is screening 30 million molecules (45 million representations) and integrating 3 different docking programs. For both projects, the consensus scores from these many millions of computations will inform the selection and downstream laboratory testing of just the top ~100 candidates, streamlining the drug development process. ======= AI-Driven Chemistry Dr. Roman Zubatyuk from the group of Prof. Olexandr Isayev at the Carnegie Mellon University chemistry department uses the access point provided by the OSG to develop a supervised machine learning method that more quickly predicts the chemical behavior of small molecules and potential drugs. Via this single access point, he is able to deploy workers globally in support of a high-throughput “Manager-worker” application. Roman reported about his OSG experience in a presentation delivered at the 2020 HTCondor Week workshop.
Astrophysics The IceCube collaboration has operated an HTCSS access point at the Wisconsin IceCube Particle Astrophysics Center for several years. It was used to access GPU capacity offered by the OSG compute federation that includes more than 20 sites of the Pacific Research Platform (PRP). As part of an <a href=”https://www.nsf.gov/awardsearch/showAward?AWD_ID=1941481&HistoricalAwards=false” target=_blank”>NSF EAGER project</a>, a team led by OSG Executive Director Frank Wuerthwein and Developer Igor Sfiligoi from the SDSC used this access point to study large-scale bursting of IceCube simulations on resources acquired from commercial cloud. In 2019, the team demonstrated a burst at the Supercomputing 19 conference that harnessed the capacity of 51K GPUs from three cloud providers and scattered across 26 cloud regions, a feat recognized as “the largest cloud simulation in history” in a keynote presentation at the conference.
Bat Genomics Dr. Ariadna Morales, used the Access Point recently deployed at the American Museum of Natural History (AMNH) to harness the capacity of local and OSG capacity in support of her work to analyze the whole genome data of 20 bats. Ariadna, a postdoc at the AMNH, used the local Access Point to run 80K compute jobs in one year. More details of her experience and other AMNH researchers can be found in the AMNH presentation at the 2020 OSG All Hands Meeting.
Gravitational Waves When the LIGO collaboration realized the need for additional capacity to analyze the observed data that led to the Nobel prize-winning detection of gravitational waves, they deployed an HTCSS Access Point that enabled them to expend their High Throughput computing capabilities to include capacity from the OSG federation and XRAC allocations. As an early adopter of HTCSS technologies, the additional capacity provided via the new Access Point easily integrated with the computing and data infrastructure of the collaboration. Deployed more than five years ago, this same access point remains an integral part of the LIGO distributed computing infrastructure, routinely integrating capacity offered by the OSG, international collaborators, XRAC allocations, and the capacity of the LIGO data grid. Read more here. (Miron acquiring link).
Drug Screening</br> The Small Molecule Screening Facility (SMSF) within UW-Madison’s Carbone Cancer Center Drug Development Core has been using an access point deployed by the UW-Madison Center for High Throughput Computing (CHTC) to perform computational docking-based screening of potential drug molecules. In just the month of November 2020, the SMSF used almost 2M core hours provided by the OSG for two projects. In collaboration with Prof. Shashank Dravid at Creighton University, one project aims to identify inhibitors of synaptic recruiting proteins for ionotropic glutamate receptors. This novel protein-protein interaction target required the docking of 14 million representations of 8 million molecules from the Aldrich chemical library using a consensus methodology developed by the SMSF to incorporate 6 different docking programs. Another project with <a href=https://apps.pharmacy.wisc.edu/sopdir/weiping_tang/index.php” target=”_blank”>Dr. Weiping Tang</a> at UW-Madison leverages the same methodology to develop novel PROTACs, that selectively degrade specific protein targets to regulate biological activity. In order to identify a ligand molecule that can recruit PROTACs to biological targets, the SMSF is screening 30 million molecules (45 million representations) and integrating 3 different docking programs. For both projects, the consensus scores from these many millions of computations will inform the selection and downstream laboratory testing of just the top ~100 candidates, streamlining the drug development process.
