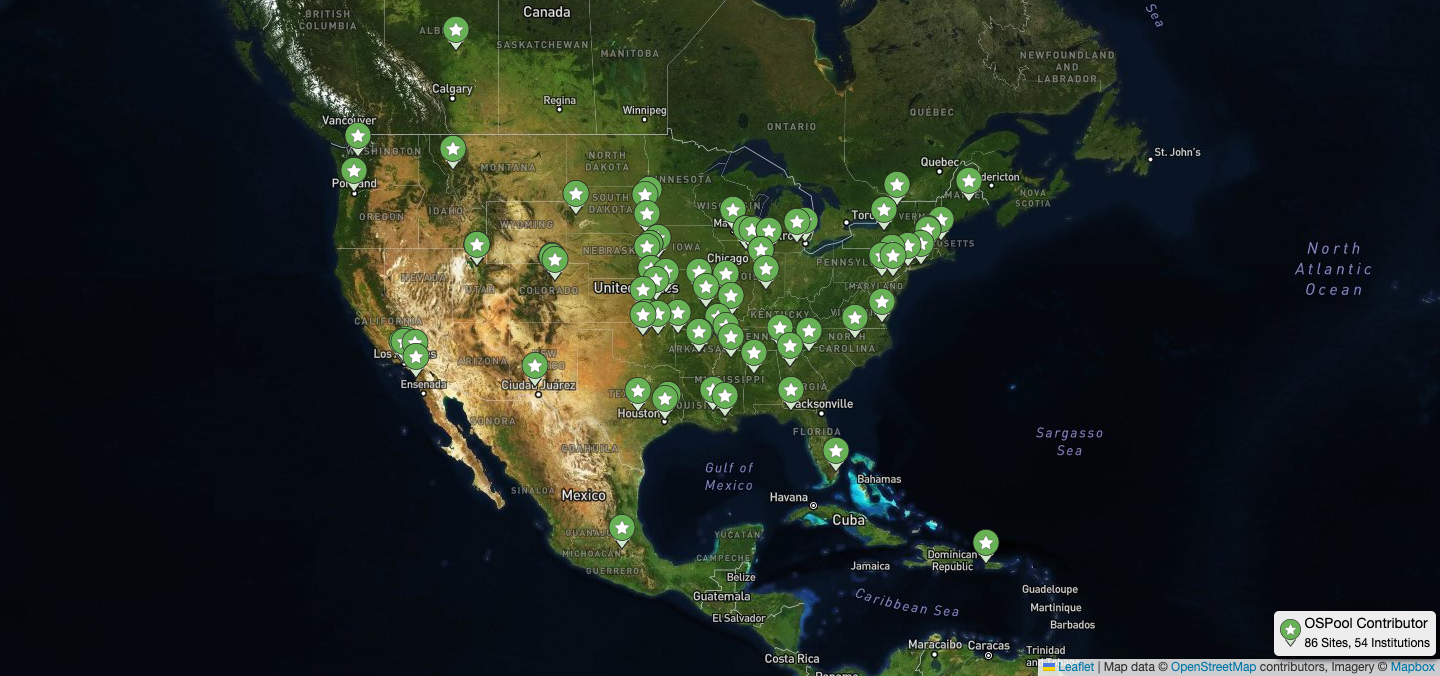
OSPool and Its Computing Capacity Helps Researchers Design New Proteins December 15, 2025
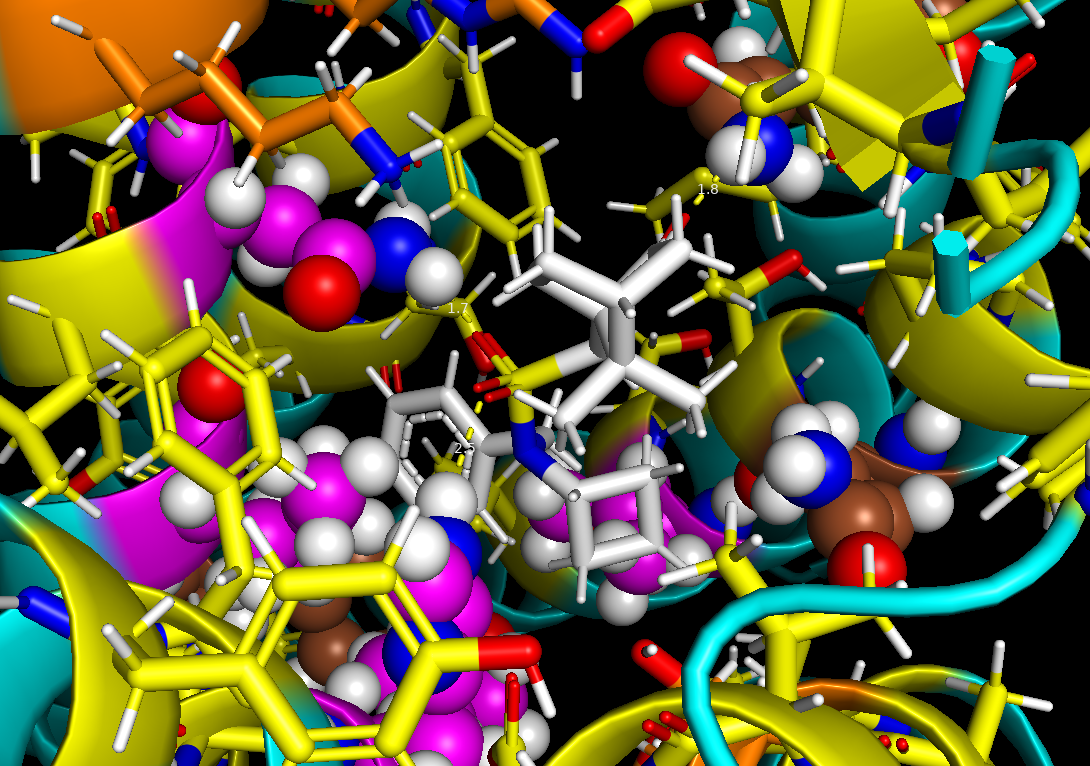
Where to find freely available GPU and CPU computing resources for research on diseases like Alzheimers, Parkinsons, and ALS? This challenge faced by labs and researchers is becoming more acute with the increasing use of AI and machine learning (ML) which demand more computing capacity. For Dr. Priyanka Joshi’s Biomolecular Homeostasis and Resilience Lab at Georgetown University, the National Science Foundation (NSF) supported OSPool and its HTCondor Software Suite was the answer.