Introducing the PATh Facility: A Unique Distributed High Throughput Computing Service
Josephine Watkins June 01, 2022
Researchers can now request credits on the PATh Facility, the PATh project’s new service intended for distributed high throughput computing workflows supporting NSF science.

With the launch of the new PATh Facility, the PATh project will soon begin providing the partnership’s first dedicated High Throughput Computing (HTC) capacity directly to researchers with NSF-funded projects. This milestone opens the door to longer runtimes, larger jobs, and greater customization for researchers. PATh is a partnership between the OSG Consortium and the University of Wisconsin-Madison’s Center for High Throughput Computing (CHTC). Jointly, the two entities have provided distributed high-throughput computing services and technologies to the S&E community for several decades.
The National Science Foundation (NSF) awards credits to access the PATh Facility, making it well-integrated in the nation’s cyberinfrastructure. Researchers can request computing credits associated with their NSF award, which they ‘cash in’ when they run HTC workloads using the PATh Facility’s services. There are currently two mechanisms to request such credit: researchers can request PATh credits within new proposals, or primary investigators (PIs) with existing awards can email their program officer to add to their award. In both cases, researchers outline the kind of HTC capacity they need; PATh’s experts are available to help researchers estimate the different requirements of their HTC workloads.
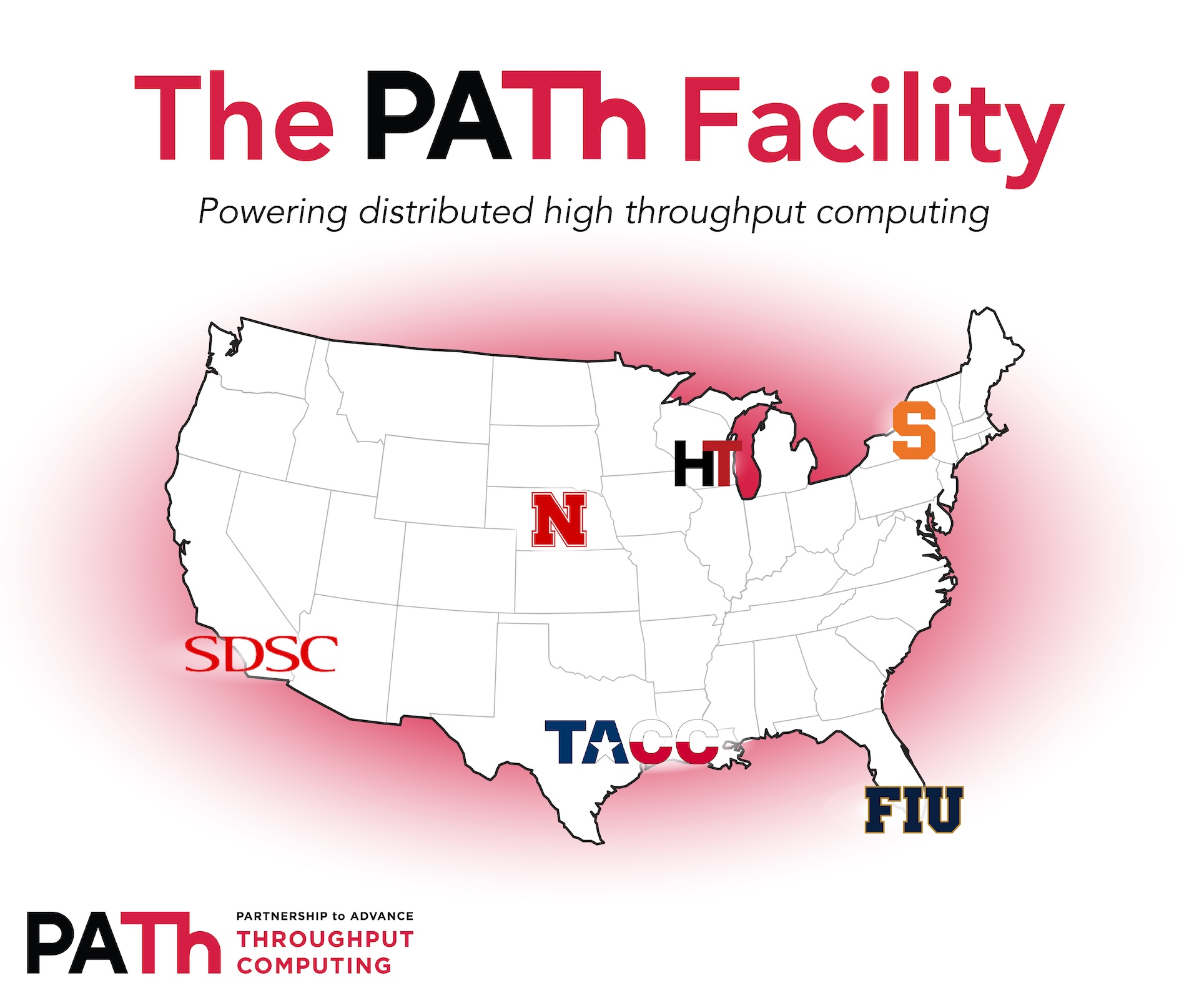
Just like the partnership, the PATh Facility is distributed and will eventually include computational resources distributed over six different sites across the nation: the Center for High Throughput Computing at the University of Wisconsin-Madison, the Holland Computing Center at the University of Nebraska-Lincoln, Syracuse University’s Research Computing group, the San Diego Supercomputing Center at University of California San Diego, the Texas Advanced Computing Center at the University of Texas at Austin, and Florida International University’s AMPATH network in Miami. This uniquely distributed resource is intended to handle HTC workloads, all for the support and advancement of NSF-funded open science. With access to the PATh Facility, researchers will have approximately 35,000 modern cores and up to 44 A100 GPUs at their fingertips.
While the PATh credit ecosystem is still growing, any PATh Facility capacity not used for credit will be available to the Open Science Pool (OSPool) to benefit all open science under a Fair-Share allocation policy. In fact, for researchers familiar with the OSPool, running HTC workloads on the PATh Facility should feel second-nature. Like the OSPool, the PATh Facility is nationally-spanning, geographically distributed, and ideal for HTC workloads. But while resources on the OSPool belong to a diverse range of campuses and organizations that have generously donated their resources to open science, the allocation of capacity in the PATh Facility is managed by the PATh Project itself.
This distinction enables longer runtimes and larger jobs otherwise infeasible on the OSPool opportunistic resources. This higher degree of control also empowers the PATh team to provide researchers with a more customized level of support. Brian Bockelman, Co-PI of the PATh Project, notes: “With the PATh Facility, we can work with researchers to come up with more bespoke solutions. Whether it’s the configuration of the hardware, the runtime, IPv6 connectivity, or whatever it is that’s not working out –– we have far more ability to change it.”
Initial facility hardware is ready for immediate use by researchers, and the remainder of the hardware is enroute to its future home. Wisconsin serves as a central hub for testing and development, and PATh Facility resources are tested there before being shipped off to their final destinations. For example, Nebraska’s share of the PATh Facility has already been shipped and is running opportunistic backfill jobs. The lights are beginning to turn on, and as Bockelman likes to say, “we’re turning electrons into science.”
However, the effort required to make the PATh Facility possible goes beyond shipping hardware and plugging in cables. To truly turn electrons into science, creativity and problem-solving will be instrumental. While the NSF is trying out new, innovative ways to award credits, PATh is responsible for credit management and tracking. This task has blossomed into an internal service development project –– the PATh development team is working on ensuring that the HTCondor Software Suite (HTCSS) can effectively track credit usage across the facility. Additionally, containers are being used as an enabling technology to provide uniform software environments across PATh Facility resources. Kubernetes, an open-source system for automating management of containerized applications, will allow PATh staff to maintain containers not just individually, but site-wide.
Marking a monumental moment for the PATh Project, the PATh Facility provides dedicated resources directly to researchers for the first time ever. The project has always been focused on advancing and democratizing access to HTC computing at all scales, and the launch of the PATh Facility makes this goal more attainable than ever. Perhaps Bockelman characterizes the facility’s impact best: “I think the unique part is the distributed aspect and the focus on high throughput computing. It extends that vision of HTC as a mechanism that can make an outsized impact on how researchers leverage computing capacity to advance their science.”
To hear more about the PATh Facility, listen to Brian Bockelman’s talk from the 2022 OSG All-Hands Meeting in March:
…
Request credits for the PATh Facility by contacting NSF. PATh Research Computing Facilitators are here to help –– please reach out to mailto:[email protected] with questions about PATh resources, using HTC, or estimating credit needs. Learn more about the PATh Facility, credit accounts, and view the 2022 Charge Listing.
